Adaptive mutations alter antibody structure and dynamics during affinity maturation
R. Adhikary, W. Yu, M. Oda, T. Chen, R. Walker, R. Stanfield, I. Wilson, J. Zimmermann, F.E. Romesberg, Biochemistry (2015) 54:2085–2093.
 In this work, three affinity matured anti-MPTS antibodies (Abs) are characterized via X-ray and spectroscopic studies and the results are discussed in terms of the elasticity, anelasticity, and plasticity of the complexes. We find that the level of plasticity of the Ab-MPTS complexes is correlated with specificity, suggesting that the optimization of protein dynamics may have contributed to affinity maturation.
In this work, three affinity matured anti-MPTS antibodies (Abs) are characterized via X-ray and spectroscopic studies and the results are discussed in terms of the elasticity, anelasticity, and plasticity of the complexes. We find that the level of plasticity of the Ab-MPTS complexes is correlated with specificity, suggesting that the optimization of protein dynamics may have contributed to affinity maturation.
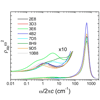 We report the sequence, thermodynamic, and time-resolved spectroscopic characterization of a panel of eight antibodies (Abs) elicited to a chromophoric antigen (MPTS). Three of the Abs arose from unique germline Abs, while the remaining five comprise two sets of siblings that arose by somatic mutation of a common precursor. By characterizing both flexibility and conformational heterogeneity, we show that point mutations are capable of fixing significant differences in protein dynamics.
We report the sequence, thermodynamic, and time-resolved spectroscopic characterization of a panel of eight antibodies (Abs) elicited to a chromophoric antigen (MPTS). Three of the Abs arose from unique germline Abs, while the remaining five comprise two sets of siblings that arose by somatic mutation of a common precursor. By characterizing both flexibility and conformational heterogeneity, we show that point mutations are capable of fixing significant differences in protein dynamics. We show that 3PEPS spectroscopy in combination with molecular dynamics simulations can provide a detailed description of protein dynamics for the fluorescein Ab 4−4−20, and how it is evolved for biological function.
We show that 3PEPS spectroscopy in combination with molecular dynamics simulations can provide a detailed description of protein dynamics for the fluorescein Ab 4−4−20, and how it is evolved for biological function. We review novel experimental methods to characterize conformational heterogeneity and dynamics in proteins.
We review novel experimental methods to characterize conformational heterogeneity and dynamics in proteins.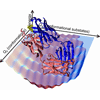 We examine a panel of antibodies elicited to the chromophoric antigen fluorescein. On the basis of isothermal titration calorimetry, we select six antibodies that bind fluorescein with diverse binding entropies, suggestive of varying contributions of dynamics to molecular recognition. We find that more than half of all the somatic mutations among the six antibodies are located in positions unlikely to contact fluorescein directly, and using 3PEPS and TG spectroscopy, we find a high level of dynamic diversity among the antibodies.
We examine a panel of antibodies elicited to the chromophoric antigen fluorescein. On the basis of isothermal titration calorimetry, we select six antibodies that bind fluorescein with diverse binding entropies, suggestive of varying contributions of dynamics to molecular recognition. We find that more than half of all the somatic mutations among the six antibodies are located in positions unlikely to contact fluorescein directly, and using 3PEPS and TG spectroscopy, we find a high level of dynamic diversity among the antibodies. We review the role of activation-induced cytidine deaminase (AID) in the complex process of somatic hypermutation (SHM) and present recent advances in experimental methods to characterize antibody dynamics as a function of SHM to help elucidate the role that the AID-induced mutations play in tailoring molecular recognition.
We review the role of activation-induced cytidine deaminase (AID) in the complex process of somatic hypermutation (SHM) and present recent advances in experimental methods to characterize antibody dynamics as a function of SHM to help elucidate the role that the AID-induced mutations play in tailoring molecular recognition. We report the characterization of protein heterogeneity and dynamics as a function of evolution for the antifluorescein antibody 4-4-20. Using nonlinear laser spectroscopy, surface plasmon resonance, and molecular dynamics simulations, we demonstrate that evolution localized the Ab-combining site from a heterogeneous ensemble of conformations to a single conformation by introducing mutations that act cooperatively and over significant distances to rigidify the protein.
We report the characterization of protein heterogeneity and dynamics as a function of evolution for the antifluorescein antibody 4-4-20. Using nonlinear laser spectroscopy, surface plasmon resonance, and molecular dynamics simulations, we demonstrate that evolution localized the Ab-combining site from a heterogeneous ensemble of conformations to a single conformation by introducing mutations that act cooperatively and over significant distances to rigidify the protein.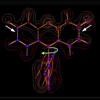 3PEPS and steady-state spectroscopy in combination with binding and structural data, are used to examine the immunological evolution of protein flexibility in an anti-fluorescein antibody. We show that antibody dynamics are systematically manipulated during affinity maturation, and suggest that the evolution of protein flexibility may be a central component of the immune response.
3PEPS and steady-state spectroscopy in combination with binding and structural data, are used to examine the immunological evolution of protein flexibility in an anti-fluorescein antibody. We show that antibody dynamics are systematically manipulated during affinity maturation, and suggest that the evolution of protein flexibility may be a central component of the immune response. We use photon echo spectroscopy to measure the response of three antibody-binding sites to perturbation from electronic excitation of a bound antigen, fluorescein. The three antibodies show motions that range in time scale from tens of femtoseconds to nanoseconds. This work was featured on the
We use photon echo spectroscopy to measure the response of three antibody-binding sites to perturbation from electronic excitation of a bound antigen, fluorescein. The three antibodies show motions that range in time scale from tens of femtoseconds to nanoseconds. This work was featured on the  The reorganization energies of the chromophore−antibody complexes are found to be distributed over a wide range. 3PEPS spectroscopy is used to measure the time scales of the reorganization process in one complex. We find that the antibody motions occur on time scales ranging from 75 femtoseconds to 67 picoseconds.
The reorganization energies of the chromophore−antibody complexes are found to be distributed over a wide range. 3PEPS spectroscopy is used to measure the time scales of the reorganization process in one complex. We find that the antibody motions occur on time scales ranging from 75 femtoseconds to 67 picoseconds.